Background
When a patient visits the hospital for medical treatment, a series of medical data is generated after diagnosis, such as disease diagnosis and surgical treatment. Medical records among different countries are similar but are written in different languages. ICD-10 is the 10th revision of the International Statistical Classification of Diseases and Related Health Problems (ICD), a medical classification list gathered by the World Health Organization (WHO). The ICD-10 code set contains codes for diseases, signs and symptoms and external causes of injury or diseases. This system provides a common language for disease classification. In the hospital, a disease coder needs an average of 20-40 minutes to classify one case. An automatic system can read free-text data such as discharge notes or history and can reduce the amount of human labor required in the hospital. The objective is described in Figure 1.

Figure 1: The objective of this research. Translation of free-text data into ICD-10 codes via a deep-learning model.
Result
This research uses word2vec to obtain a word vector to measure syntactic and semantic word similarities. Created by a team of researchers led by Tomas Mikolov at Google, word2vec is a group of related models used to produce word embeddings. Word2vec [1], a two-layer neural network, uses free-text data as input to construct a vector space. In this space, each unique word in the input corpus obtains a corresponding word vector. Word vectors are located in the position where they share common context in the user’s input free-text data. Thus, the word vectors can be used to train the neural network for classification. In the neural network model, the input is the word vector trained from the free-text medical data about the patient's situation. The output is the ICD-10 codes corresponding to the patient's disease. Our model uses a convolutional recurrent neural network as the model architecture. The embedding dimension is 100, which means that each word is replaced by a 100-dimensional vector, and the loss function is categorical cross entropy. Adam is the optimizer for neural network training. The model architecture is described in Figure 2. This model can obtain an f-measure of approximately 0.9 for the 22-categorical classification.
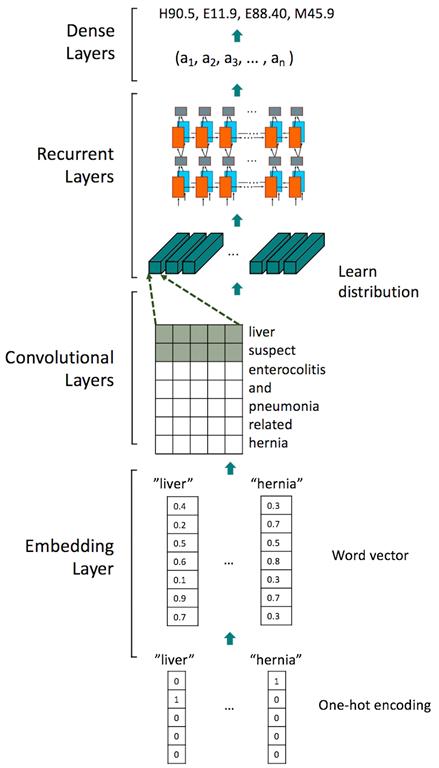
Figure 2: The model architecture in this research.
This model can provide disease coders hints in classification work to help them with the first few codes and thereby speed up their classification. The proposed method can classify the first three digits with an f-measure of 0.7, and the goal of future work is to improve the results sufficiently to replace disease coders. By using word2vec and the neural network, computers can understand free-text data that can only be read by humans. Computer can learn the semantics underlying the language and help humans perform the otherwise laborious work. In this the proposed method, the input data are written in English. However, word2vec can transform all types of language into word vectors. Thus, hospitals in other countries can use this method to classify their free-text medical data using ICD-10 codes as well. Certainly, we can change the input data into other free-text medical data such as gene description and the output data into genes. A model for classifying genes based on free-text medical data about genes can thus be obtained.
Reference
Mikolov, T., Sutskever, I., Chen, K., Corrado, G., & Dean, J. (2013). Distributed Representations of Words and Phrases and their Compositionality. In NIPS'13 Proceedings of the 26th International Conference on Neural Information Processing Systems, 2, 3111-3119.
Feipei Lai
Professor, Graduate Institute of Biomedical Electronics and Bioinformatics, the Department of Computer Science & Information Engineering and the Department of Electrical Engineering
Yu-Hsuan Chang
Institute of Biomedical Electronics and Bioinformatics