The World Health Organization has predicted that
the prevalence of bronchial asthma will increase globally in forthcoming years.
Early life gut microbial dysbiosis is currently recognized as a key determinant
of immune dysregulation, which is associated with a broad spectrum of allergic
disorders. However, we still do not know what or how gut microbe(s) are
responsible for allergy establishment. It is arguable that early and rapid
colonization by Clostridium/Bacteroides/Escherichia
coli and insufficient colonization by Lactobacilli/Bifidobacterium
contribute to allergy development in infants.1-6 The controversy of
this topic is most likely due to the use of different assay systems as well as
the fact that traditional bacterial cultivation methods normally produce lower
than expected numbers of detectable species.4, 5, 7 Additionally,
many relevant studies have been cross-sectional and have not included serial
sampling.1,4-6 Some studies have enrolled cases with current atopic
diseases and lacked sufficient data on the microbiota configurations that
existed before disease onset.2, 3 The control groups in these studies
deserve the most critical appraisal as the studies have usually enrolled
control participants from different families unrelated to the allergic patients;1-8
as a result, the genetic and environmental factors that modulate microbial
colonization could not be precisely controlled. All of these drawbacks restrict
the efficient and accurate identification of the bacteria that predisposed
individuals to the development of allergies, and this represents a missed
chance to achieve successful treatment.
We assumed that dysbiosis of the gut microbiota in infancy may predispose patients to later allergy development, and our intention in this study was to identify the causative bacteria. The aforementioned design drawbacks were avoided in this study because we enrolled a twin cohort to diminish the biases caused by familial and environmental determinants. We collected the twins’ stool samples serially beginning on the first day of life to allow an ample time window for the early identification of related microbial patterns before disease development. Once a pair of twins exhibited discordant clinical phenotypes, their fecal collections were compared, and we began to attempt to tease out whether this is a culprit in the disease. We employed culture-free PCR-temporal temperature gradient gel electrophoresis (TTGE) and 16S rRNA-based next-generation sequencing (NGS) to profile the entire microbiome down to the species level. By using this comprehensive and rigorous strategy, we provide the first evidence indicating that is the relevant bacteria that causes allergic diseases. This bacterial species was rarely found in non-allergic subjects but was present at a significantly higher frequency prior to or concurrent with the onset of allergic manifestations.
To confirm this bedside finding in a bench study, we established an in vivo asthmatic mouse model as well as an in vitro coculture system of bacteria and murine colon tissues to corroborate the role of R. gnavus in allergic manifestations. The immunoregulatory mechanisms underlying the competence of R. gnavus to control allergy development were studied not only in lung tissues but also in the colon, where the bacteria colonized. R. gnavus has been demonstrated to be a mucolytic bacterium capable of degrading colonic mucin and consuming mucin-derived glycans.9 The data presented in this study data further demonstrate that after it penetrated the mucus layer, R. gnavus was recognized by dendritic cells (DCs), and the colonic epithelia were stimulated to secrete epithelium-derived cytokines like IL-33. These cytokines elicited an expansion in type 2 innate lymphoid cells (ILC2s) and DCs to prime the differentiation of Th2 cells from naïve T cells, thus ensuring the production of Th2 cytokines and causing tissue eosinophilic infiltration. As a result, Th2 lymphocytes and effector cells rapidly circulated to the lung via the circulatory and lymphatic systems to initiate an asthmatic inflammatory cascade.
In conclusion, we have exposed a potential microbiota-driven gut-lung axis that represents a mechanism by which dysbiosis could cause allergic diseases in infants. Our data indicate that R. gnavus-associated dysbiosis induced Th2-biased immunity in the colon and also exploited the gut-pulmonary axis to evoke allergic asthma, thus constituting a potential therapeutic target for the treatment and prevention of the disease.
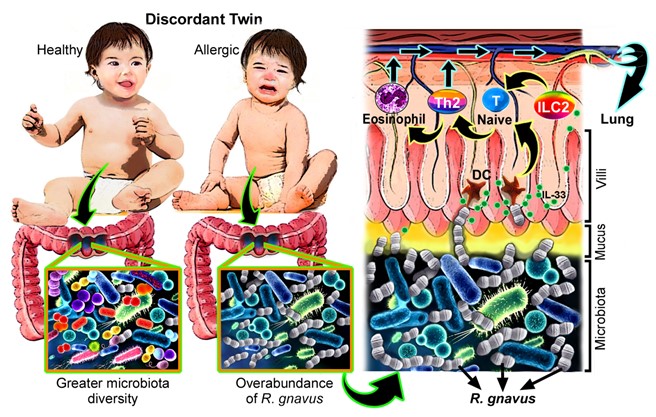
Figure 1. Difference in gut microbiota configurations between allergic discordant twins. Dysbiosis characterized by a low diversity of gut microbiota but an increased number of R. gnavus was frequently found in diseased twins, whereas healthy twins tended to have greater microbiota diversity. Prior to inducing Th2 dominance in allergic twins, R. gnavus penetrates the colonic mucus layer and stimulates the underlying colonic epithelium to secrete epithelium-derived cytokines such as IL-33. Simultaneously, this bacterium is recognized by DCs, resulting in the activation of DCs and ILC2s, which promote the differentiation of Th2 cells and consequential colonic infiltration by eosinophils. These effectors enter the circulatory/lymphatic systems along with Th2 cells before moving to the lung to initiate the manifestations of asthma.
References
1. Penders J, Thijs C, van den Brandt PA, et al. (2007). Gut microbiota composition and development of atopic manifestations in infancy: the KOALA Birth Cohort Study. Gut, 56, 661-667. DOI: 10.1136/gut.2006.100164
2. Candela M, Rampelli S, Turroni S, et al. (2012). Unbalance of intestinal microbiota in atopic children. BMC Microbiol, 12, 95-104. DOI: 10.1186/1471-2180-12-95.
3. Gore C, Munro K, Lay C, et al. (2008). Bifidobacterium pseudocatenulatum is associated with atopic eczema: a nested case-control study investigating the fecal microbiota of infants. J Allergy Clin Immunol, 121, 135-140. DOI: 10.1016/j.jaci.2007.07.061.
4. Vael C, Nelen V, Verhulst SL, et al. (2008). Early intestinal Bacteroides fragilis colonisation and development of asthma. BMC Pulm Med, 8, 19-25. DOI: 10.1186/1471-2466-8-19.
5. Kendler M, Uter W, Rueffer A, et al. (2006). Comparison of fecal microflora in children with atopic eczema/dermatitis syndrome according to IgE sensitization to food. Pediatr Allergy Immunol, 17, 141-147. DOI: 10.1111/j.1399-3038.2005.00371.x.
6. Wang M, Karlsson C, Olsson C, et al. (2008). Reduced diversity in the early fecal microbiota of infants with atopic eczema. J Allergy Clin Immunol, 121, 129-134. DOI: 10.1016/j.jaci.2007.09.011.
7. Adlerberth I, Strachan DP, Matricardi PM, et al. (2007). Gut microbiota and development of atopic eczema in 3 European birth cohorts. J Allergy Clin Immunol, 120, 343-350. DOI: 10.1016/j.jaci.2007.05.018.
8. Storro O, Oien T, Langsrud O, et al. (2011). Temporal variations in early gut microbial colonization are associated with allergen-specific immunoglobulin E but not atopic eczema at 2 years of age. Clin Exp Allergy, 41, 1545-1554. DOI: 10.1016/j.jaci.2007.05.018.
9. Crost EH, Tailford LE, Le Gall G, et al. (2013). Utilisation of mucin glycans by the human gut symbiont Ruminococcus gnavus is strain-dependent. PLoS One, 8, e76341. DOI: 10.1371/journal.pone.0076341.
Yen-Hsuan Ni,
Department of Pediatrics, School of Medicine