Secondary cell wall (SCW) biosynthesis is the biological process that generates wood, an important renewable feedstock for materials and energy. NAC domain transcription factors, particularly Vascular- Related NAC-Domain (VND) and Secondary Wall-Associated NAC Domain (SND) proteins, are known to regulate SCW differentiation. In plants, the regulation of VND and SND is important to maintain homeostasis to avoid abnormal growth and development. We previously identified PtrSND1-A2IR, a splice variant derived from PtrSND1-A2 that acts as a dominant-negative regulator, which suppresses the transactivation of all PtrSND1 family members. PtrSND1-A2IR also suppresses the self-activation of the PtrSND1 family members except for its cognate transcription factor, PtrSND1-A2, suggesting the existence of an unknown factor needed to regulate PtrSND1-A2. Here, PtrVND6-C1IR, a splice variant derived from PtrVND6-C1, was discovered; this variant suppresses the protein functions of all PtrVND6 family members. PtrVND6-C1IR also suppresses the expression of all PtrSND1 members, including PtrSND1-A2, demonstrating that PtrVND6-C1IR is the previously unidentified regulator of PtrSND1-A2. We also found that PtrVND6-C1IR cannot suppress the expression of its cognate transcription factor, PtrVND6-C1. PtrVND6-C1 is suppressed by PtrSND1-A2IR. Neither PtrVND6-C1IR nor PtrSND1-A2IR can suppress its cognate transcription factors, but both variants can suppress all members of the other family. The results indicate that the splice variants from the PtrVND6 and PtrSND1 families may exert reciprocal cross-regulation to produce complete transcriptional regulation of these two families during wood formation. This reciprocal cross-regulation between families suggests that an additional level of regulation provided by intron-retained splice variants may be a general regulatory mechanism among NAC domain proteins and likely for other transcription factors.
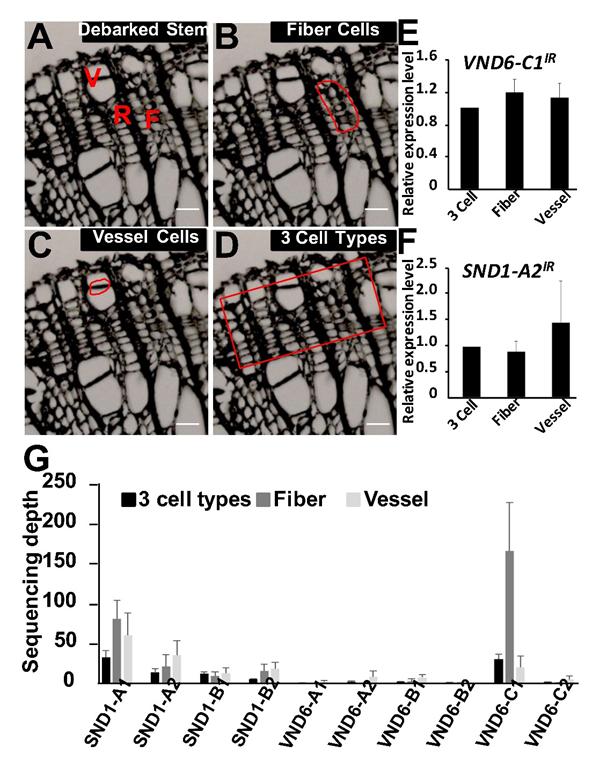
Figure 1. Fiber cells, vessel cells, and three cell types collected using LCM and transcript abundances of the PtrVND6 and PtrSND1 families in these cell types. (A) Cross-section of the debarked stem from the 17th internode of P. trichocarpa. F, fiber cells; R, ray cells; V, vessel cells. LCM was used to collect (B) fiber cells, (C) vessel cells, and (D) three cell types (scale bars, 25 µm). qRT-PCR was used to detect the transcript abundance of (E) PtrVND6-C1IR and (F) PtrSND1-A2IR in different cell types. (G) RNA-seq analysis was used to estimate the transcript abundances of full-length PtrVND6 and PtrSND1. The error bars represent the SE from three biological replicates.
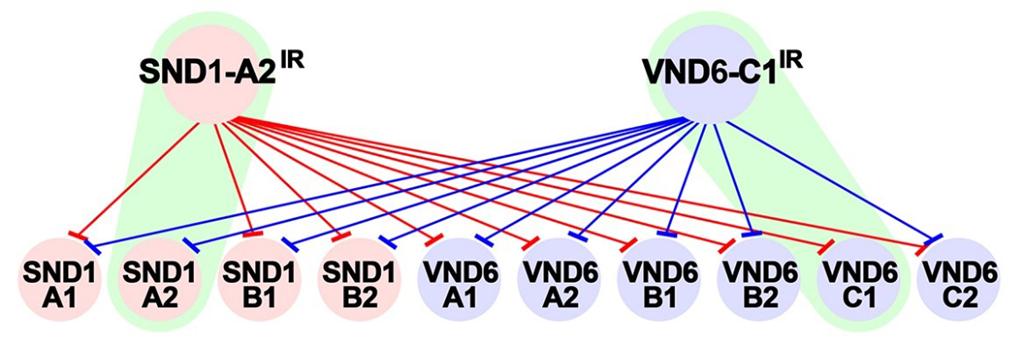
Figure 2. Cross-regulation between the PtrVND6 and PtrSND1 families through PtrVND6-C1IR and PtrSND1-A2IR. PtrSND1-A2IR suppresses the transcript abundance of the PtrSND1 and PtrVND6 families (red edges) except PtrSND1-A2 (green highlight). PtrVND6-C1IR suppresses PtrSND1 and PtrVND6 families (blue edges) except PtrVND6-C1 (green highlight).
Reference
Lin, Y. J., Chen, H., Li, Q., Li, W., Wang, J. P., Shi, R., Tunlaya-Anukit, S., Shuai, P., Wang, Z., Ma, H., Li, H., Sun, Y., Sederoff, R. R., Chiang, V. L. (2017). Reciprocal cross-regulation of VND and SND multi-gene TF families in wood formation in Populus trichocarpa. Proceedings of the National Academy of Sciences of the United States of America (PNAS), 114(45), E9722–E9729. DOI:10.1073/pnas.1714422114.
Ying-Chung Jimmy Lin
Assistant Professor, Department of Life Sciences
Assistant Professor (joint appointment), Institute of Plant Biology